Introduction
This simple tutorial demonstrates how to get CogStack running on a sample electronic health record (EHR) dataset stored initially in an external database. CogStack ecosystem has been designed with handling efficiently both structured and unstructured EHR data in mind. It shows its strength while working with the unstructured type of data, especially as some input data can be provided as documents in PDF or image formats. For the moment, however, we only show how to run CogStack on a set of structured and free-text EHRs that have been already digitalized. The part covering unstructured type of data in form of PDF documents, images and other clinical notes which needs to processed prior to analysis will come shortly.
This tutorial is divided into 5 parts:
- Getting CogStack (link),
- A brief description of how does CogStack work and its ecosystem (link),
- A brief description of the sample datasets used (link),
- Running CogStack ‘out-of-the-box’ using the dataset already preloaded into a sample database (link),
- For advanced users: preparing a database schema according to the sample dataset and to the current CogStack pipeline requirements (link),
- For advanced users: preparing the configuration file for CogStack data processing engine according to the used database schema and used microservices (link).
To skip the brief description and to get hands on running CogStack please head directly to Running CogStack part.
The main directory with resources used in this tutorial is available in the CogStack bundle under examples/. This tutorial is based on the Example 2, however, there are more examples available to play with.
Getting CogStack
The most convenient way to get CogStack bundle is to download it directly from the official github repository either by cloning it using git:
git clone -b dev --single-branch https://github.com/CogStack/CogStack-Pipeline.git
or by downloading it from the repository and decompressing it:
wget 'https://github.com/CogStack/CogStack-Pipeline/archive/dev.zip'
unzip dev.zip
The content will be decompressed into CogStack-Pipeline/ directory.
Note: For the moment the CogStack bundle is obtained from the dev branch – soon it will be merged into master branch with a version tag for a direct download.
How does CogStack work
Data processing workflow
The data processing workflow of CogStack is based on Java Spring Batch framework. Not to dwell too much into technical details and just to give a general idea – the data is being read from a predefined data source, later it follows a number of processing operations with the final result stored in a predefined data sink. CogStack implements variety of data processors, data readers and writers with scalability mechanisms that can be selected in CogStack job configuration. Although the data can be possibly read from different sources, the most frequently used data sink is ElasicSearch. For more details about the CogStack functionality, please refer to the CogStack Documentation (WIP).
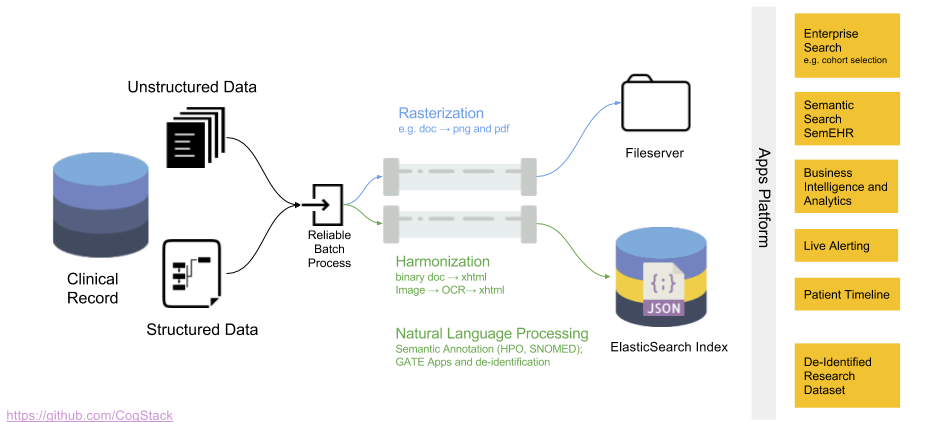
In this tutorial we only focus on a simple and very common use-case, where CogStack reads and process structured and free-text EHRs data from a single PostgreSQL database. The result is then stored in ElasticSearch where the data can be easily queried in Kibana dashboard. However, CogStack data processing engine also supports multiple data sources – please see Example 3 which covers such case.
CogStack ecosystem
CogStack ecosystem consists of multiple inter-connected microservices running together. For the ease of use and deployment we use Docker (more specifically, Docker Compose), and provide Compose files for configuring and running the microservices. The selection of running microservices depends mostly on the specification of EHR data source(s), data extraction and processing requirements.
In this tutorial the CogStack ecosystem is composed of the following microservices:
pgsamples– PostgreSQL database loaded with a sample dataset underdb_samplesname,cogstack– CogStack data processing pipeline with worker(s),postgres– PostgreSQL database for storing information about CogStack jobs,elasticsearch– ElasticSearch search engine (single node) for storing and querying the processed EHR data,kibana– Kibana data visualization tool for querying the data from ElasticSearch.
The Docker Compose file with configuration of these microservices can be found in examples/example2/docker/docker-compose.yml.
Sample datasets
The sample dataset used in this tutorial consists of two types of EHR data:
- Synthetic – structured, synthetic EHRs, generated using synthea application,
- Medial reports – unstructured, medical health report documents obtained from MTsamples.
These datasets, although unrelated, are used together to compose a combined dataset.
Synthetic – synthea-based
This dataset consists of synthetic EHRs that were generated using synthea application – the synthetic patient generator that models the medical history of generated patients. For this tutorial, we generated EHRs for 100 patients and exported them as CSV files. Typed in the main synthea directory, the command line for running it:
./run_synthea -p 100 \
--generate.append_numbers_to_person_names=false \
--exporter.csv.export=true
However, the pre-generated files are provided in a compressed form as examples/rawdata/synsamples.tgz file.
The generated dataset consists of the following files:
allergies.csv– Patient allergy data,careplans.csv– Patient care plan data, including goals,conditions.csv– Patient conditions or diagnoses,encounters.csv– Patient encounter data,imaging_studies.csv– Patient imaging metadata,immunizations.csv– Patient immunization data,medications.csv– Patient medication data,observations.csv– Patient observations including vital signs and lab reports,patients.csv– Patient demographic data,procedures.csv– Patient procedure data including surgeries.
For more details about the generated files and the schema definition please refer to the official synthea wiki page. The sample records are shown in Advanced: preparing a DB schema for CogStack part.
In this example we use a subset of the available data – as a simple use-case, only patients.csv, encounters.csv and observations.csv file are used related. These files also represent separate tables in the db_samples database. For more advanced use-cases please check the Example 3 which uses the full dataset.
Medical reports – MTSamples
MTsamples is a collection of transcribed medical sample reports and examples. The reports are in a free-text format and have been downloaded directly from the official website.
Each report contain such information as:
- Sample Type,
- Medical Specialty,
- Sample Name,
- Short Description,
- Medical Transcription Sample Report (in free text format).
The collection comprises in total of 4873 documents. A sample document is shown in Advanced: preparing a DB schema for CogStack part.
Preparing the data
For the ease of use a database dump with predefined schema and preloaded data will be provided in examples/example2/db_dump directory. This way, the PostgreSQL database will be automatically initialized when deployed using Docker. The database dump for this example (alongside the others) can be also directly downloaded from Amazon S3 by running in the main examples/ directory:
bash download_db_dumps.sh
Alternatively, the PostgreSQL database schema definition used in this tutorial db_create_schema.sql is stored in examples/example2/extra/ directory alongside the script prepare_db.sh to generate the database dump. More information covering the creation of database schema can be found in Advanced: preparing a DB schema for CogStack part.
Running CogStack
Running CogStack for the first time
For the ease of use CogStack is being deployed and run using Docker. However, before starting the CogStack ecosystem for the first time, one needs to have the database dump files for sample data either by creating them locally or downloading from Amazon S3.
To download the database dumps, just type in the main examples/ directory:
bash download_db_dumps.sh
Next, a setup scripts needs to be run locally to prepare the Docker images and configuration files for CogStack data processing pipeline. The script is available in examples/example2/ path and can be run as:
bash setup.sh
As a result, a temporary directory __deploy/ will be created containing all the necessary artifacts to deploy CogStack.
Docker-based deployment
Next, we can proceed to deploy CogStack ecosystem using Docker Compose. It will configure and start microservices based on the provided Compose file: examples/example2/docker/docker-compose.yml. Moreover, the PostgreSQL database container comes with pre-initialized database dump ready to be loaded directly into. In order to run CogStack, type in the examples/example2/__deploy/ directory:
docker-compose up
In the console there will be printed status logs of the currently running microservices. For the moment, however, they may be not very informative (we’re working on that).
Connecting to the microservices
CogStack ecosystem
The picture below sketches a general idea on how the microservices are running and communicating within a sample CogStack ecosystem used in this tutorial.
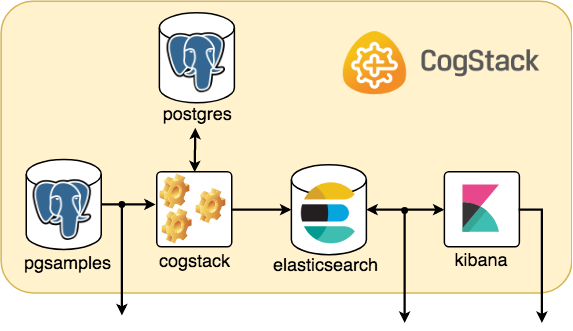
Assuming that everything is working fine, we should be able to connect to the running microservices. Selected running services (elasticsearch and kibana) have their port connections forwarded to host localhost. When accessing webservices and when asked for credentials the username is test with password test.
Kibana and ElasticSearch
Kibana dashboard used to query the EHRs can be accessed directly in browser via URL: http://localhost:5601/. The data can be queried using a number of ElasticSearch indices, e.g. sample_observations_view. Usually, each index will correspond to the database view in db_samples (pgsamples PostgreSQL database) from which the data was ingested. However, when entering Kibana dashboard for the first time, an index pattern needs to be configured in the Kibana management panel – for more information about its creation, please refer to the official Kibana documentation.
In addition, ElasticSearch REST end-point can be accessed via URL http://localhost:9200/. It can be used to perform manual queries or to be used by other external services – for example, one can list the available indices:
curl 'http://localhost:9200/_cat/indices'
or query one of the available indices – sample_observations_view:
curl 'http://localhost:9200/sample_observations_view'
For more information about possible documents querying or modification operations, please refer to the official ElasticSearch documentation.
PostgreSQL sample database
Moreover, the access PostgreSQL database with the input sample data is exposed directly at localhost:5555. The database name is db_sample with user test and password test. To connect, one can run:
psql -U 'test' -W -d 'db_samples' -h localhost -p 5555
The information about connecting to the micro-services and resources will become useful in Advanced: preparing a configuration file for CogStack part.
Advanced: preparing a DB schema for CogStack
General information
In the current implementation, CogStack can only ingest EHR data from a specified input database. This is why, in order to process the sample patient data covered in this tutorial, one needs to create an appropriate database schema and load the data.
Moreover, as relational join statements have a high performance burden for ElasticSearch, the EHR data is best to be stored denormalized in ElasticSearch. This is why, for the moment, we rely on ingesting the data from additional view(s) created in the sample database.
Following, we cover the process of defining the required schema step-by-step.
Database schema – tables
Patients table
The first 5 records of patient data (file: patients.csv from Synthea-based samples) in CSV format is presented below:
ID,BIRTHDATE,DEATHDATE,SSN,DRIVERS,PASSPORT,PREFIX,FIRST,LAST,SUFFIX,MAIDEN,MARITAL,RACE,ETHNICITY,GENDER,BIRTHPLACE,ADDRESS,CITY,STATE,ZIP
b9f5a11b-211d-4ced-b3ba-12012c83b937,1939-08-04,1996-03-15,999-11-9633,S99999830,X106007X,Mr.,Brady,Lynch,,,M,white,polish,M,Worcester,701 Schiller Esplanade,Fitchburg,Massachusetts,01420
fab43860-c3be-4808-b7b4-00423c02816b,1962-06-21,2011-03-10,999-67-8307,S99958025,X26840237X,Mrs.,Antonia,Benavides,,Padrón,M,hispanic,mexican,F,Rockland,643 Hand Bay,Boston,Massachusetts,02108
84dd6378-2ddc-44b6-9292-2a4461bcef53,1998-12-01,,999-50-5147,S99987241,,Mr.,Keith,Conn,,,,white,english,M,Rockland,461 Spinka Extension Suite 69,Framingham,Massachusetts,01701
9929044f-1f43-4453-b2c0-a2f45dcdd4be,2014-09-23,,999-64-4171,,,,Derrick,Lakin,,,,white,irish,M,Tewksbury,577 Hessel Lane,Hampden,Massachusetts,
The patients table definition in PostgreSQL according to the specification:
create table patients (
ID uuid primary key,
BIRTHDATE date,
DEATHDATE date,
SSN varchar(64),
DRIVERS varchar(64),
PASSPORT varchar(64),
PREFIX varchar(8),
FIRST varchar(64),
LAST varchar(64),
SUFFIX varchar(8),
MAIDEN varchar(64),
MARITAL char(1),
RACE varchar(64),
ETHNICITY varchar(64),
GENDER char(1),
BIRTHPLACE varchar(64),
ADDRESS varchar(64),
CITY varchar(64),
STATE varchar(64),
ZIP varchar(64)
) ;
Encounters table
Similarly, the first 5 records of patient encounters data (file: encounters.csv)
ID,START,STOP,PATIENT,CODE,DESCRIPTION,COST,REASONCODE,REASONDESCRIPTION
123ffd84-618e-47cd-abca-5fe95b72179a,1955-07-30T07:30Z,1955-07-30T07:45Z,b9f5a11b-211d-4ced-b3ba-12012c83b937,185345009,Encounter for symptom,129.16,36971009,Sinusitis (disorder)
66b016e9-e797-446a-8f2b-e5534acbbb04,1962-03-09T07:30Z,1962-03-09T07:45Z,b9f5a11b-211d-4ced-b3ba-12012c83b937,185349003,Encounter for check up (procedure),129.16,,
d517437d-50b5-4cab-aca2-9c010c06989e,1983-08-19T07:30Z,1983-08-19T08:00Z,b9f5a11b-211d-4ced-b3ba-12012c83b937,185349003,Encounter for check up (procedure),129.16,,
2452cf09-021b-4586-9e33-59d7d2242f31,1987-08-28T07:30Z,1987-08-28T07:45Z,b9f5a11b-211d-4ced-b3ba-12012c83b937,185349003,Encounter for check up (procedure),129.16,,
f25a828f-ae79-4dd0-b6eb-bca26138421b,1969-09-13T14:02Z,1969-09-13T14:34Z,fab43860-c3be-4808-b7b4-00423c02816b,185345009,Encounter for symptom,129.16,10509002,Acute bronchitis (disorder)
with the corresponding encounters table definition:
create table encounters (
ID uuid primary key,
START timestamp,
STOP timestamp,
PATIENT uuid references patients,
CODE varchar(64),
DESCRIPTION text,
COST real,
REASONCODE varchar(64),
REASONDESCRIPTION text,
DOCUMENT text -- (*)
) ;
Here, with --(*) has been marked an additional DOCUMENT field. This extra field will be used to store a document from MTSamples dataset.
Just to clarify, Synthea-based and MTSamples are two unrelated datasets. Here, we are extending the synthetic dataset with the clinical documents from the MTSamples to create a combined one, to be able to perform more advanced queries.
A sample document from MTSamples dataset is presented below:
Sample Type / Medical Specialty: Allergy / Immunology
Sample Name: Allergic Rhinitis
Description: A 23-year-old white female presents with complaint of allergies.
(Medical Transcription Sample Report)
SUBJECTIVE: This 23-year-old white female presents with complaint of allergies. She used to have allergies when she lived in Seattle but she thinks they are worse here. In the past, she has tried Claritin, and Zyrtec. Both worked for short time but then seemed to lose effectiveness. She has used Allegra also. She used that last summer and she began using it again two weeks ago. It does not appear to be working very well. She has used over-the-counter sprays but no prescription nasal sprays. She does have asthma but doest not require daily medication for this and does not think it is flaring up.
MEDICATIONS: Her only medication currently is Ortho Tri-Cyclen and the Allegra.
ALLERGIES: She has no known medicine allergies.
OBJECTIVE:
Vitals: Weight was 130 pounds and blood pressure 124/78.
HEENT: Her throat was mildly erythematous without exudate. Nasal mucosa was erythematous and swollen. Only clear drainage was seen. TMs were clear.
Neck: Supple without adenopathy.
Lungs: Clear.
ASSESSMENT: Allergic rhinitis.
PLAN:
1. She will try Zyrtec instead of Allegra again. Another option will be to use loratadine. She does not think she has prescription coverage so that might be cheaper.
2. Samples of Nasonex two sprays in each nostril given for three weeks. A prescription was written as well.
Keywords: allergy / immunology, allergic rhinitis, allergies, asthma, nasal sprays, rhinitis, nasal, erythematous, allegra, sprays, allergic,
Observations table
The next table is observations, where the first 5 rows of observations.csv file are:
DATE,PATIENT,ENCOUNTER,CODE,DESCRIPTION,VALUE,UNITS,TYPE
1987-08-28,b9f5a11b-211d-4ced-b3ba-12012c83b937,2452cf09-021b-4586-9e33-59d7d2242f31,8302-2,Body Height,180.3,cm,numeric
2002-06-27,fab43860-c3be-4808-b7b4-00423c02816b,fd113dfd-5e2c-40f2-98f3-e153665c3f53,8302-2,Body Height,165.2,cm,numeric
2009-04-07,84dd6378-2ddc-44b6-9292-2a4461bcef53,2058da3b-f7f3-4ebc-8c1e-360cd256cdcb,8302-2,Body Height,138.3,cm,numeric
2002-06-27,fab43860-c3be-4808-b7b4-00423c02816b,fd113dfd-5e2c-40f2-98f3-e153665c3f53,29463-7,Body Weight,81.8,kg,numeric
2009-04-07,84dd6378-2ddc-44b6-9292-2a4461bcef53,2058da3b-f7f3-4ebc-8c1e-360cd256cdcb,29463-7,Body Weight,24.6,kg,numeric
and the corresponding table definition:
create table observations (
CID serial primary key, -- (*)
DCT timestamp default current_timestamp, -- (*)
DATE date,
PATIENT uuid references patients,
ENCOUNTER uuid references encounters,
CODE varchar(64),
DESCRIPTION text,
VALUE varchar(64),
UNITS varchar(64),
TYPE varchar(64),
) ;
Here, with --(*) have been marked additional fields with auto-generated values. These are: CID – an automatically generated primary key and DCT – a document creation timestamp. They will be later used by CogStack pipeline for data partitioning when processing the records. The patient and encouters tables have their primary keys (ID field) already defined (of uuid type) and are included in the input CSV files.
Database schema – views
Next, we define a observations_view that will be used by CogStack data processing engine to ingest the records from input database:
create view observations_view as
select
p.ID as patient_id,
p.BIRTHDATE as patient_birth_date,
p.DEATHDATE as death_date,
p.SSN as patient_SSN,
p.DRIVERS as patient_drivers,
p.PASSPORT as patient_passport,
p.PREFIX as patient_prefix,
p.FIRST as patient_first_name,
p.LAST as patient_last_name,
p.SUFFIX as patient_suffix,
p.MAIDEN as patient_maiden,
p.MARITAL as patient_marital,
p.RACE as patient_race,
p.ETHNICITY as patient_ethnicity,
p.GENDER as patient_gender,
p.BIRTHPLACE as patient_birthplace,
p.ADDRESS as patient_addr,
p.CITY as patient_city,
p.STATE as patient_state,
p.ZIP as patient_zip,
enc.ID as encounter_id,
enc.START as encounter_start,
enc.STOP as encounter_stop,
enc.CODE as encounter_code,
enc.DESCRIPTION as encounter_desc,
enc.COST as encounter_cost,
enc.REASONCODE as encounter_reason_code,
enc.REASONDESCRIPTION as encounter_reason_desc,
enc.DOCUMENT as encounter_document,
obs.DATE as observation_date,
obs.CODE as observation_code,
obs.DESCRIPTION as observation_desc,
obs.VALUE as observation_value,
obs.UNITS as observation_units,
obs.TYPE as observation_type,
-- for CogStack compatibility
'src_field_name'::text as cog_src_field_name, -- (a)
'observations_view'::text as cog_src_table_name, -- (b)
obs.CID as cog_pk, -- (c)
'cog_pk'::text as cog_pk_field_name, -- (d)
obs.DCT as cog_update_time -- (e)
from
patients p,
encounters enc,
observations obs
where
enc.PATIENT = p.ID and
obs.PATIENT = p.ID and
obs.ENCOUNTER = enc.ID
;
The goal here is to denormalize the database schema for CogStack and ElasticSearch data ingestion, as the observations table is referencing both the patient and encounters tables by their primary key. In the current implementation, CogStack engine cannot yet perform dynamic joins over the relational data from specific database tables.
Apart from exposing the fields from the previously defined tables, some extra fields cog_* have been added. They are required for compatibility with CogStack data processing engine, but they may be possibly removed or modified in the upcoming version of CogStack. However, in the current implementation, these fields are required to properly configure the CogStack database reader, database row mapper, to properly schedule and partition the data of the running CogStack data processing workers.
These additional fields are:
cog_src_field_name– related with processing the text documents (not used in this example),cog_src_table_name– the name of the table containing records to process,cog_pk– primary key value (or any unique) used for partitioning the data into batches (for the moment, needs to be of numeric type),cog_pk_field_name– the name of the field in the current table/view containing the value of primary key values,cog_update_time– the last update/modification time of the record, used for checking for new records and for partitioning.
These fields are later used when preparing the configuration file for CogStack data processing workflow.
Advanced: preparing a configuration file for CogStack
General information
Each CogStack data processing pipeline is configured using a number of parameters defined in the corresponding Java properties file. Moreover, multiple CogStack pipelines can be launched in parallel (see Example 3), each using its own properties file with configuration. In this example we use only one pipeline with configuration specified in examples/example2/cogstack/observations.properties file.
Properties description
There are multiple configurable parameters available to tailor the CogStack data processing pipeline to the specific data processing needs and available resources. Here we will cover only the most important parameters related with configuring the input source, the output sink and data processing workflow. For a more detailed description of the available parameters, please refer to the CogStack documentation (WIP).
Spring profiles
CogStack configuration file uses Spring profiles, which enable different components of the data processing pipeline. In our example we use:
spring.profiles.active = jdbc_in,elasticsearchRest,localPartitioning
which denotes that only such profiles will be active:
jdbc_infor JDBC input database connector,elasticsearchRestfor using REST API for inserting documents to ElasticSearch,partitioningfunctionality.
For a more detailed description of available profiles please refer to CogStack documentation.
Data source
The parameters for specifying the data source are defined as follows:
source.JdbcPath = jdbc:postgresql://pgsamples:5432/db_samples
source.Driver = org.postgresql.Driver
source.username = test
source.password = test
source.poolSize = 10
In this example we are using a PostgreSQL database which driver is defined by source.Driver parameter. The PostgreSQL database service is available in the CogStack ecosystem as pgsamples, has exposed port 5432 and the sample database name is db_samples – all these details need to be included in the source.JdbcPath parameter field. The information about the data source host and port directly corresponds to the pgsamples microservice configuration specified in the Docker Compose file (examples/example2/docker/docker-compose.yml) as mentioned in the Running CogStack part.
Please note that the source.poolSize defines the maximum size of the connection pool available for performing queries by CogStack workers. A PostgreSQL database, by default, has a maximum connection limit set to 100, hence exceeding the limit (either by a single job or multiple parallel ones) may lead to termination of the data pipeline.
Next, we need to instruct CogStack workers how to query the records from the data source:
source.selectClause = SELECT *
source.sortKey = cog_pk
source.fromClause = FROM observations_view
source.srcTableName = cog_src_table_name
source.srcColumnFieldName = cog_src_field_name
source.primaryKeyFieldName = cog_pk_field_name
source.primaryKeyFieldValue = cog_pk
source.timeStamp = cog_update_time
source.dbmsToJavaSqlTimestampType = TIMESTAMP
This is where the previously defined observations_view with additional CogStack-specific fields are used (see: Advanced: preparing a DB schema for CogStack part).
Data sink
Next, we need to define the data sink – in our example, and by default, ElasticSearch is being used:
elasticsearch.cluster.name = elasticsearch
elasticsearch.cluster.host = elasticsearch
elasticsearch.cluster.port = 9200
elasticsearch.security.enabled = false
elasticsearch.ssl.enabled = false
Similarly, as when defining the sample database source, we need to provide the ElasticSearch host and port configuration according to the microservices definition in the corresponding Docker Compose file.
As an additional feature, security and ssl encryption can be enabled for communication with ElasticSearch. However, it uses the ElasticSearch X-Pack bundle and requires license for commercial deployments, hence it is disabled by default.
In the next step, we specify the ElasticSearch indexing parameters:
elasticsearch.index.name = sample_observations_view
elasticsearch.type = doc
elasticsearch.excludeFromIndexing = cog_pk,cog_pk_field_name,cog_src_field_name,cog_src_table_name
elasticsearch.datePattern = yyyy-MM-dd'T'HH:mm:ss.SSS
We specify the index name which will be used to store the documents processed by CogStack workers. Additionally, we specify which fields should be excluded from the indexing – by default, we exclude the binary content, the constant-value fields and the primary key from the observations_view (see: Advanced: preparing a DB schema for CogStack).
As a side note, although it’s mostly for testing purposes at the moment, the same information regarding the source database host and credentials needs to be provided for defining the target (in our case it is ElasticSearch anyway):
target.JdbcPath = jdbc:postgresql://pgsamples:5432/db_samples
target.Driver = org.postgresql.Driver
target.username = test
target.password = test
This is a concept that will be possibly redesigned in future versions of CogStack.
Jobs and CogStack workers configuration
CogStack engine in order to coordinate the workers needs to keep the information about the current jobs in an additional PostgreSQL database – postgres. Hence, similarly as when defining the source database, this database needs to specified:
jobRepository.JdbcPath = jdbc:postgresql://postgres:5432/cogstack
jobRepository.Driver = org.postgresql.Driver
jobRepository.username = cogstack
jobRepository.password = mysecretpassword
job.jobName = job_observations_view
The last parameter job.jobName is a default name for the jobs that will be created.
Partitioner and scheduler
Another set of useful parameters are related with controlling the job execution and data partitioning:
partitioner.partitionType = PKTimeStamp
partitioner.tableToPartition = observations_view
partitioner.pkColumnName = cog_pk
partitioner.timeStampColumnName = cog_update_time
In the current implementation, CogStack engine can only partition the data using the records’ primary key (cog_pk field, containing unique values) and records’ update time (cog_update_time field) as defined in observations_view. This is specified by PKTimeStamp partitioning method types.
Apart from data partitioning, it can be useful to set up the scheduler – the following line corresponds to the scheduler configuration:
scheduler.useScheduling = false
In this example we do not use scheduler, since we ingest EHRs from the data source only once. However, in case when the data is being generated in a continuous way, scheduler should be enabled to periodically run CogStack jobs to process the new EHRs.